Dr Mark Nanyingi is a postdoctoral research associate at the School of Public Health at the University of Nairobi and holds an honorary appointment at the Institute of Infection and Global Health, University of Liverpool. He is an infectious disease epidemiologist with a focus on One Health and modelling of zoonotic diseases.
Globally, zoonotic diseases continue to impact negatively on public health, the livestock sector and economies. In Kenya, there have been multisectoral efforts to generate information on zoonoses through evidence-based research that in turn supports and guides the government’s prevention and control strategies. For the last decade, Dr Nanyingi has been working closely with the Zoonotic Disease Unit (ZDU), which is Kenya’s One Health coordinating office between the ministries of health and livestock. In 2015, as part of a multi-disciplinary team of experts, Dr Nanyingi used using a semi-quantitative One Health Zoonotic Disease Prioritization tool developed by the Centres for Disease Control and Prevention to prioritise 36 zoonotic diseases based on severity of illness in humans, epidemic potential, socio-economic burden, prevalence/incidence and availability of interventions. The top five Priority Zoonotic Diseases (PZD) were Anthrax, Trypanosomiasis, Rabies, Brucellosis and Rift Valley fever (Munyua et al., 2016). Under the HORN project, Dr Nanyingi is engaged in operational research and development of strategic frameworks for some of these diseases, and most recently has focused on Rift Valley Fever.
Rift Valley fever (RVF) is a vector borne, viral zoonosis that causes significant economic and public health impacts in animals and humans. Kenya has experienced several RVF outbreaks since 1931, the most notable ones being the 1997–1998 and 2006–2007 outbreaks. The occurrence of the disease is associated with heavy rainfall and flooding which provides ideal conditions for mosquito vector breeding, multiplication and disease emergence. Mosquitoes of the Aedes species are the primary vectors, while the Culex and Anopheles species and other biting flies are secondary, amplifying vectors that propagate transmission.
In June 2018, a national outbreak of RVF was declared by the Kenya Directorate of Veterinary Services, following reports of abortions and death of young sheep, camels and goats in Wajir County, located in the North Eastern part of the country. Four human deaths were reported from Wajir, characterised by febrile and haemorrhagic illness, and another human death was reported in Siaya County, in Western Kenya. All of these were confirmed by laboratory analysis at the Kenya Medical Research Institute, Nairobi.
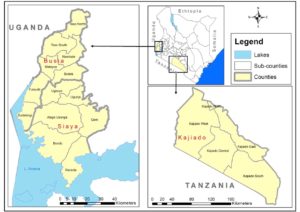
Dr Nanyingi’s current research focuses on establishing an integrated RVF One Health Surveillance system in three low endemic areas of Kenya as part of the government’s decision to support tools that will lead to faster disease detection, cost effective prevention and targeted control strategies of RVF during the interepidemic period (Oyas et al 2018). The areas of study will are Siaya, Busia and Kajiado. Busia was selected based on evidence from previous studies that documented presence of RVF in humans and animals in the Lake Victoria crescent of western Kenya (Cook et al., 2017, Grossi-Soyster et al, 2019).
This is human-animal linked cross-sectional study. Objectives include determining the seroprevalence of RVF infection in ruminants (cattle, sheep and goats) and seroprevalence and risk factors for seropositivity among febrile patients. The study further intends to characterize the distribution and diversity of potential RVF vectors while evaluating the molecular diversity of RVF Virus (RVFV) strains circulating in Kenya. Using the serological evidence we will develop spatial predictive models that will contribute to the existing risk maps for RVF in Kenya.
Using hospital-based prospective and multicentre cross-sectional surveys, febrile patients attending selected health facilities will be targeted, their clinical history will be obtained and physical examination performed on consenting patients. Blood will be collected for serological determination of RVF virus by molecular techniques. Similarly, Livestock will be sampled in the same study areas by cluster techniques, where the primary sampling unit will be a village, which comprises of several homesteads. The sampling will ensure even distribution of villages that should be within at least 10 km radius of the previous epidemic foci or seropositive areas. Blood will be collected and serum samples will be tested for antiRVF IgG antibodies. Whole Genome sequencing of the animal and human samples will be conducted to construct phylogenetic trees that may explain genetic ancestry of circulating strains with the previous outbreaks.
This study aims to identify serological evidence of RVF in humans and animals in low endemic areas that may demonstrate the subclinical circulation of RVFV. Complete genome sequences of the RVFV human and livestock strains will contribute to the understanding of the dynamics of RVFV evolution and adaptation in Kenya, and RVF risk maps and models will predict the potential areas of transmission of RVFV in Kenya.

